Not all DNA play by
the same rules!
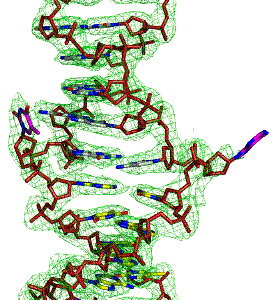
Our group explores the dynamic world of nucleic acids beyond the familiar B-DNA double helix, exploring how non-canonical DNA and RNA structures influence gene regulation-from transcription to translation and everything in between.
We investigate how these structures are formed, what stabilizes them, and how they interact with cellular machinery to impact diseases. We have established the chemical-induced formation of B-Z junctions with base extrusion to better understand their structural and cellular implications.
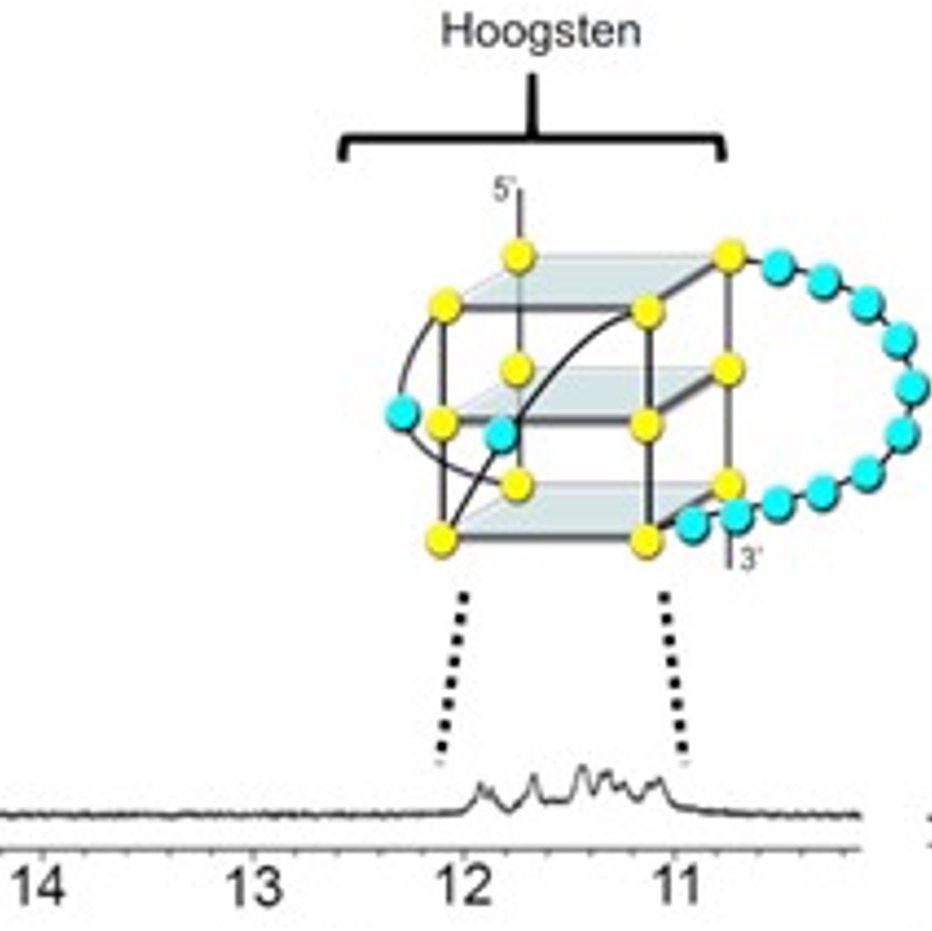

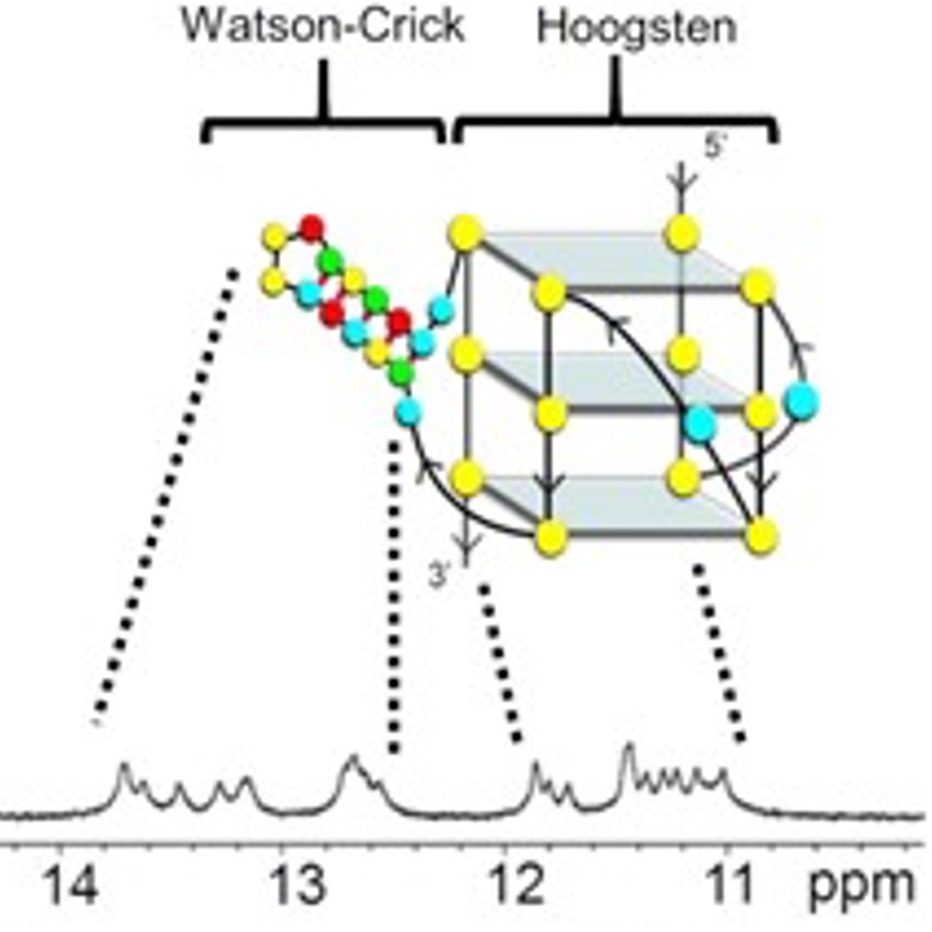
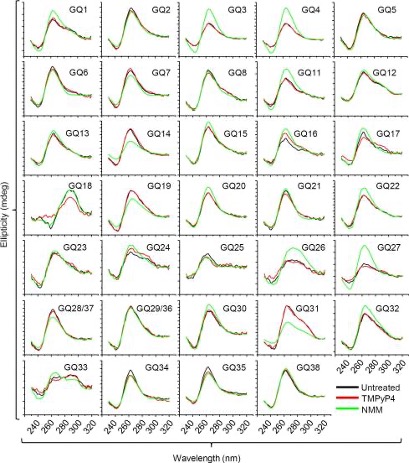
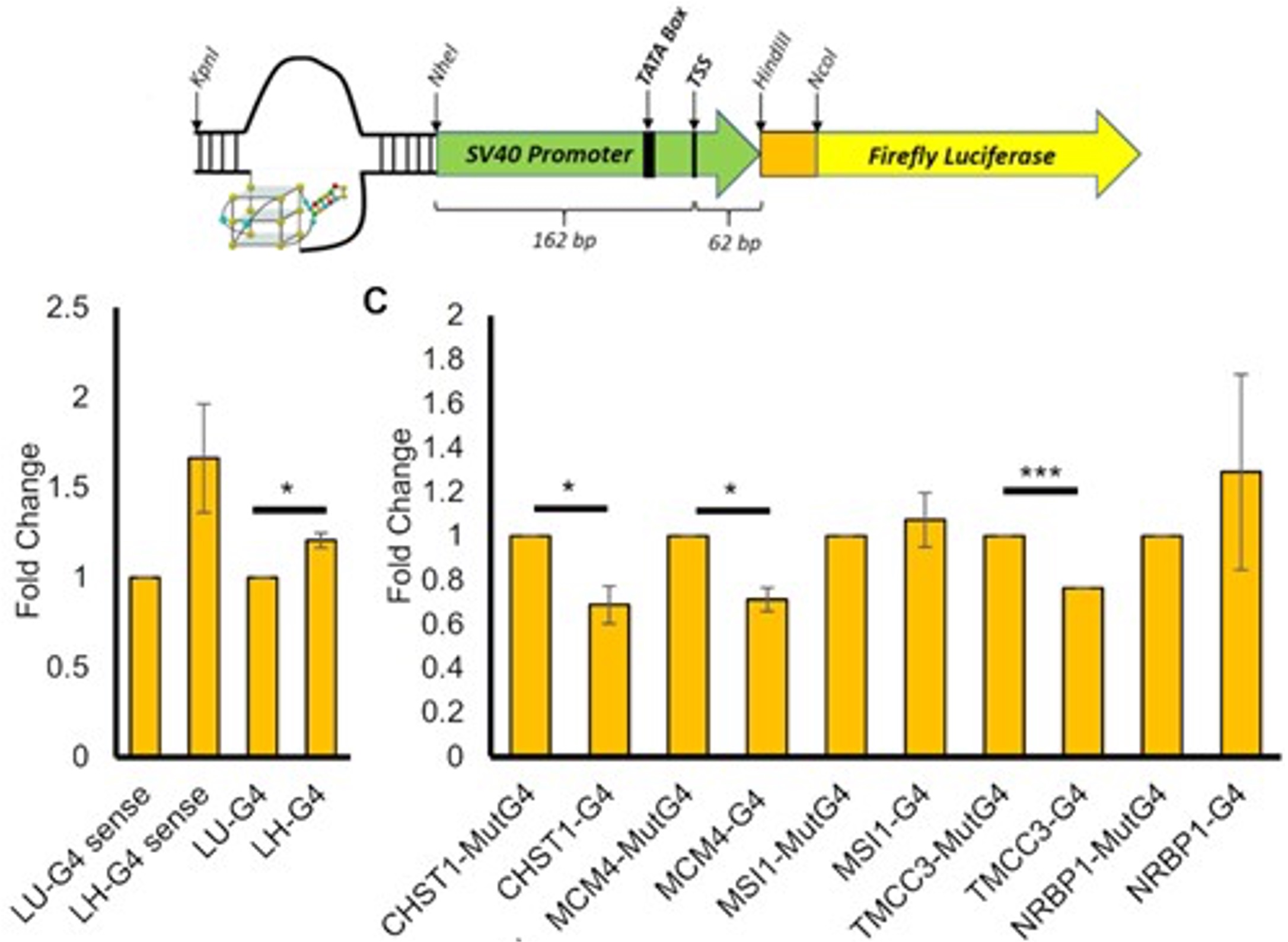
Our group also places strong emphasis on G-quadruplexes (G4s), investigating how they influence genetic regulation in both cancer and viral systems. Our work has unraveled a G4-dependent mechanism of transcriptional regulation, demonstrating significant enrichment at oncogenic promoter, where the formation of transcriptional condensates and promoter activation are highly dependent on G4-mediated protein binding. We identified G-quadruplexes as unique and promising druggable targets in human cytomegalovirus and SARS-CoV infections. Our on-going work expands G4-targeting strategies across additional viral systems and cancer models.

We are developing new molecular approaches that influence alternative nucleic acid conformations and build versatile platforms that seek to turn the structural “oddities” of the genome into new therapeutic opportunities.
Recent Publications
1. Yoo, W., Song, Y.W., Bansal, V. and Kim, K.K., 2025. G-quadruplex-dependent transcriptional regulation by molecular condensation in the Bcl3 promoter. Nucleic Acids Research, 53(16), p.gkaf827.
2. Sultan, M., Razzaq, M., Lee, J., Das, S., Kannappan, S., Subramani, V.K., Yoo, W., Kim, T., Lee, H.R., Chaurasia, A.K. and Kim, K.K., 2025. Targeting the G-quadruplex as a novel strategy for developing antibiotics against hypervirulent drug-resistant Staphylococcus aureus. Journal of Biomedical Science, 32(1), p.15.
3. Kannappan, S., Jo, K., Kim, K.K. and Lee, J.H., 2024. Utilizing peptide-anchored DNA templates for novel programmable nanoparticle assemblies in biological macromolecules: A review. International Journal of Biological Macromolecules, 256, p.128427.
4. Yoo, W., Song, Y.W., Kim, J., Ahn, J., Kim, J., Shin, Y., Ryu, J.K. and Kim, K.K., 2023. Molecular basis for SOX2-dependent regulation of super-enhancer activity. Nucleic Acids Research, 51(22), pp.11999-12019.
5. Razzaq, M., Han, J.H., Ravichandran, S., Kim, J., Bae, J.Y., Park, M.S., Kannappan, S., Chung, W.C., Ahn, J.H., Song, M.J. and Kim, K.K., 2023. Stabilization of RNA G-quadruplexes in the SARS-CoV-2 genome inhibits viral infection via translational suppression. Archives of pharmacal research, 46(7), pp.598-615.
6. Subramani, V.K. and Kim, K.K., 2023. Characterization of Z-DNA using circular dichroism. In Z-DNA: Methods and Protocols (pp. 33-51). New York, NY: Springer US.
7. Ravichandran, S., Razzaq, M., Parveen, N., Ghosh, A. and Kim, K.K., 2021. The effect of hairpin loop on the structure and gene expression activity of the long-loop G-quadruplex. Nucleic acids research, 49(18), pp.10689-10706.
8. Hur, J.H., Kang, C.Y., Lee, S., Parveen, N., Yu, J., Shamim, A., Yoo, W., Ghosh, A., Bae, S., Park, C.J. and Kim, K.K., 2021. AC-motif: a DNA motif containing adenine and cytosine repeat plays a role in gene regulation. Nucleic Acids Research, 49(17), pp.10150-10165.
9. Shamim, A., Razzaq, M. and Kim, K.K., 2020. MD-TSPC4: computational method for predicting the thermal stability of I-motif. International Journal of Molecular Sciences, 22(1), p.61.